Bar plot of gene expression for 1-, 2-, or 3-factor experiments
Source:R/plotFactor.r
plotFactor.RdCreates a bar plot of relative gene expression (fold change) values from 1-, 2-, or 3-factor experiments, including error bars and statistical significance annotations.
Usage
plotFactor(
data,
x_col,
y_col,
Lower.se_col,
Upper.se_col,
group_col = NULL,
facet_col = NULL,
letters_col = NULL,
letters_d = 0.2,
col_width = 0.8,
err_width = 0.15,
dodge_width = 0.8,
fill_colors = NULL,
alpha = 1,
base_size = 12,
legend_position = "right",
...
)Arguments
- data
Data frame containing expression results
- x_col
Character. Column name for x-axis
- y_col
Character. Column name for bar height
- Lower.se_col
Character. Column name for lower SE
- Upper.se_col
Character. Column name for upper SE
- group_col
Character. Column name for grouping bars (optional)
- facet_col
Character. Column name for faceting (optional)
- letters_col
Character. Column name for significance letters (optional)
- letters_d
Numeric. Vertical offset for letters (default
0.2)- col_width
Numeric. Width of bars (default
0.8)- err_width
Numeric. Width of error bars (default
0.15)- dodge_width
Numeric. Width of dodge for grouped bars (default
0.8)- fill_colors
Optional vector of fill colors
- alpha
Numeric. Transparency of bars (default
1)- base_size
Numeric. Base font size for theme (default
12)- legend_position
Character or numeric vector. Legend position (default
right)- ...
Additional ggplot2 layer arguments
Examples
data <- read.csv(system.file("extdata", "data_2factorBlock.csv", package = "rtpcr"))
res <- ANOVA_DCt(data,
numOfFactors = 2,
block = "block",
numberOfrefGenes = 1)
#> NULL
#>
#> Relative expression (DCt method)
#> factor1 factor2 RE log2FC LCL UCL se Lower.se.RE Upper.se.RE
#> 1 S L2 2.9545 1.5629 4.2644 2.0470 0.0551 2.8438 3.0695
#> 2 R L2 0.9837 -0.0238 1.4198 0.6815 0.0841 0.9280 1.0427
#> 3 S L0 0.7916 -0.3371 1.1426 0.5485 0.2128 0.6831 0.9175
#> 4 R L1 0.6240 -0.6804 0.9006 0.4323 0.4388 0.4603 0.8458
#> 5 S L1 0.4126 -1.2771 0.5956 0.2859 0.2540 0.3460 0.4921
#> 6 R L0 0.2838 -1.8171 0.4096 0.1966 0.0208 0.2797 0.2879
#> Lower.se.log2FC Upper.se.log2FC sig
#> 1 1.5044 1.6237 a
#> 2 -0.0252 -0.0224 b
#> 3 -0.3907 -0.2908 b
#> 4 -0.9223 -0.5020 bc
#> 5 -1.5230 -1.0709 cd
#> 6 -1.8435 -1.7911 d
#>
#> Combined Expression Table (all genes)
#> gene factor1 factor2 RE log2FC LCL UCL se Lower.se.RE
#> 1 PO S L2 2.9545 1.5629 4.2644 2.0470 0.0551 2.8438
#> 2 PO R L2 0.9837 -0.0238 1.4198 0.6815 0.0841 0.9280
#> 3 PO S L0 0.7916 -0.3371 1.1426 0.5485 0.2128 0.6831
#> 4 PO R L1 0.6240 -0.6804 0.9006 0.4323 0.4388 0.4603
#> 5 PO S L1 0.4126 -1.2771 0.5956 0.2859 0.2540 0.3460
#> 6 PO R L0 0.2838 -1.8171 0.4096 0.1966 0.0208 0.2797
#> Upper.se.RE Lower.se.log2FC Upper.se.log2FC sig
#> 1 3.0695 1.5044 1.6237 a
#> 2 1.0427 -0.0252 -0.0224 b
#> 3 0.9175 -0.3907 -0.2908 b
#> 4 0.8458 -0.9223 -0.5020 bc
#> 5 0.4921 -1.5230 -1.0709 cd
#> 6 0.2879 -1.8435 -1.7911 d
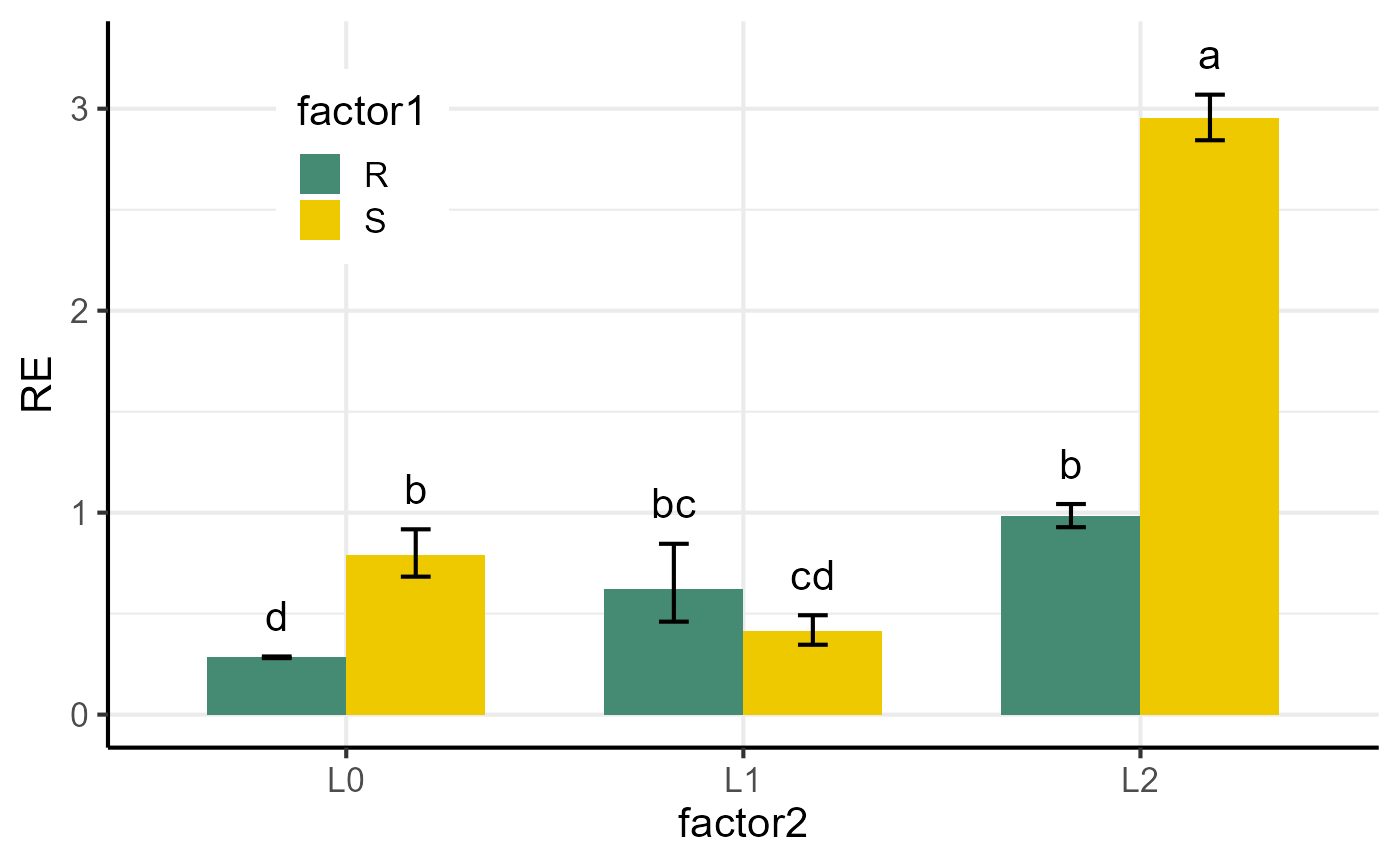
df <- res$combinedResults
p1 <- plotFactor(
data = df,
x_col = "factor2",
y_col = "RE",
group_col = "factor1",
Lower.se_col = "Lower.se.RE",
Upper.se_col = "Upper.se.RE",
letters_col = "sig",
letters_d = 0.2,
fill_colors = c("aquamarine4", "gold2"),
alpha = 1,
col_width = 0.7,
dodge_width = 0.7,
base_size = 16,
legend_position = c(0.2, 0.8))
p1
 data <- read.csv(system.file("extdata", "data_3factor.csv", package = "rtpcr"))
#Perform analysis first
res <- ANOVA_DCt(
data,
numOfFactors = 3,
numberOfrefGenes = 1,
block = NULL)
#> NULL
#>
#> Relative expression (DCt method)
#> Type Conc SA RE log2FC LCL UCL se Lower.se.RE Upper.se.RE
#> 1 S H A2 5.1934 2.3767 8.1197 3.3217 0.1309 4.7428 5.6867
#> 2 S H A1 2.9690 1.5700 4.6420 1.8990 0.0551 2.8578 3.0846
#> 3 R H A2 1.7371 0.7967 2.7159 1.1110 0.0837 1.6391 1.8409
#> 4 S L A2 1.5333 0.6167 2.3973 0.9807 0.0865 1.4441 1.6280
#> 5 R H A1 0.9885 -0.0167 1.5455 0.6323 0.0841 0.9325 1.0479
#> 6 S L A1 0.7955 -0.3300 1.2438 0.5088 0.2128 0.6864 0.9220
#> 7 S M A2 0.7955 -0.3300 1.2438 0.5088 0.2571 0.6657 0.9507
#> 8 R M A1 0.6271 -0.6733 0.9804 0.4011 0.4388 0.4626 0.8500
#> 9 S M A1 0.4147 -1.2700 0.6483 0.2652 0.2540 0.3477 0.4945
#> 10 R M A2 0.3150 -1.6667 0.4925 0.2015 0.2890 0.2578 0.3848
#> 11 R L A1 0.2852 -1.8100 0.4459 0.1824 0.0208 0.2811 0.2893
#> 12 R L A2 0.0641 -3.9633 0.1002 0.0410 0.8228 0.0362 0.1134
#> Lower.se.log2FC Upper.se.log2FC sig
#> 1 2.1705 2.6025 a
#> 2 1.5112 1.6311 ab
#> 3 0.7517 0.8443 bc
#> 4 0.5808 0.6548 c
#> 5 -0.0177 -0.0157 cd
#> 6 -0.3825 -0.2847 d
#> 7 -0.3944 -0.2761 d
#> 8 -0.9127 -0.4968 de
#> 9 -1.5145 -1.0650 ef
#> 10 -2.0363 -1.3641 f
#> 11 -1.8363 -1.7841 f
#> 12 -7.0103 -2.2407 g
#>
#> Combined Expression Table (all genes)
#> gene Type Conc SA RE log2FC LCL UCL se Lower.se.RE
#> 1 PO S H A2 5.1934 2.3767 8.1197 3.3217 0.1309 4.7428
#> 2 PO S H A1 2.9690 1.5700 4.6420 1.8990 0.0551 2.8578
#> 3 PO R H A2 1.7371 0.7967 2.7159 1.1110 0.0837 1.6391
#> 4 PO S L A2 1.5333 0.6167 2.3973 0.9807 0.0865 1.4441
#> 5 PO R H A1 0.9885 -0.0167 1.5455 0.6323 0.0841 0.9325
#> 6 PO S L A1 0.7955 -0.3300 1.2438 0.5088 0.2128 0.6864
#> 7 PO S M A2 0.7955 -0.3300 1.2438 0.5088 0.2571 0.6657
#> 8 PO R M A1 0.6271 -0.6733 0.9804 0.4011 0.4388 0.4626
#> 9 PO S M A1 0.4147 -1.2700 0.6483 0.2652 0.2540 0.3477
#> 10 PO R M A2 0.3150 -1.6667 0.4925 0.2015 0.2890 0.2578
#> 11 PO R L A1 0.2852 -1.8100 0.4459 0.1824 0.0208 0.2811
#> 12 PO R L A2 0.0641 -3.9633 0.1002 0.0410 0.8228 0.0362
#> Upper.se.RE Lower.se.log2FC Upper.se.log2FC sig
#> 1 5.6867 2.1705 2.6025 a
#> 2 3.0846 1.5112 1.6311 ab
#> 3 1.8409 0.7517 0.8443 bc
#> 4 1.6280 0.5808 0.6548 c
#> 5 1.0479 -0.0177 -0.0157 cd
#> 6 0.9220 -0.3825 -0.2847 d
#> 7 0.9507 -0.3944 -0.2761 d
#> 8 0.8500 -0.9127 -0.4968 de
#> 9 0.4945 -1.5145 -1.0650 ef
#> 10 0.3848 -2.0363 -1.3641 f
#> 11 0.2893 -1.8363 -1.7841 f
#> 12 0.1134 -7.0103 -2.2407 g
df <- res$combinedResults
df
#> gene Type Conc SA RE log2FC LCL UCL se Lower.se.RE
#> 1 PO S H A2 5.1934 2.3767 8.1197 3.3217 0.1309 4.7428
#> 2 PO S H A1 2.9690 1.5700 4.6420 1.8990 0.0551 2.8578
#> 3 PO R H A2 1.7371 0.7967 2.7159 1.1110 0.0837 1.6391
#> 4 PO S L A2 1.5333 0.6167 2.3973 0.9807 0.0865 1.4441
#> 5 PO R H A1 0.9885 -0.0167 1.5455 0.6323 0.0841 0.9325
#> 6 PO S L A1 0.7955 -0.3300 1.2438 0.5088 0.2128 0.6864
#> 7 PO S M A2 0.7955 -0.3300 1.2438 0.5088 0.2571 0.6657
#> 8 PO R M A1 0.6271 -0.6733 0.9804 0.4011 0.4388 0.4626
#> 9 PO S M A1 0.4147 -1.2700 0.6483 0.2652 0.2540 0.3477
#> 10 PO R M A2 0.3150 -1.6667 0.4925 0.2015 0.2890 0.2578
#> 11 PO R L A1 0.2852 -1.8100 0.4459 0.1824 0.0208 0.2811
#> 12 PO R L A2 0.0641 -3.9633 0.1002 0.0410 0.8228 0.0362
#> Upper.se.RE Lower.se.log2FC Upper.se.log2FC sig
#> 1 5.6867 2.1705 2.6025 a
#> 2 3.0846 1.5112 1.6311 ab
#> 3 1.8409 0.7517 0.8443 bc
#> 4 1.6280 0.5808 0.6548 c
#> 5 1.0479 -0.0177 -0.0157 cd
#> 6 0.9220 -0.3825 -0.2847 d
#> 7 0.9507 -0.3944 -0.2761 d
#> 8 0.8500 -0.9127 -0.4968 de
#> 9 0.4945 -1.5145 -1.0650 ef
#> 10 0.3848 -2.0363 -1.3641 f
#> 11 0.2893 -1.8363 -1.7841 f
#> 12 0.1134 -7.0103 -2.2407 g
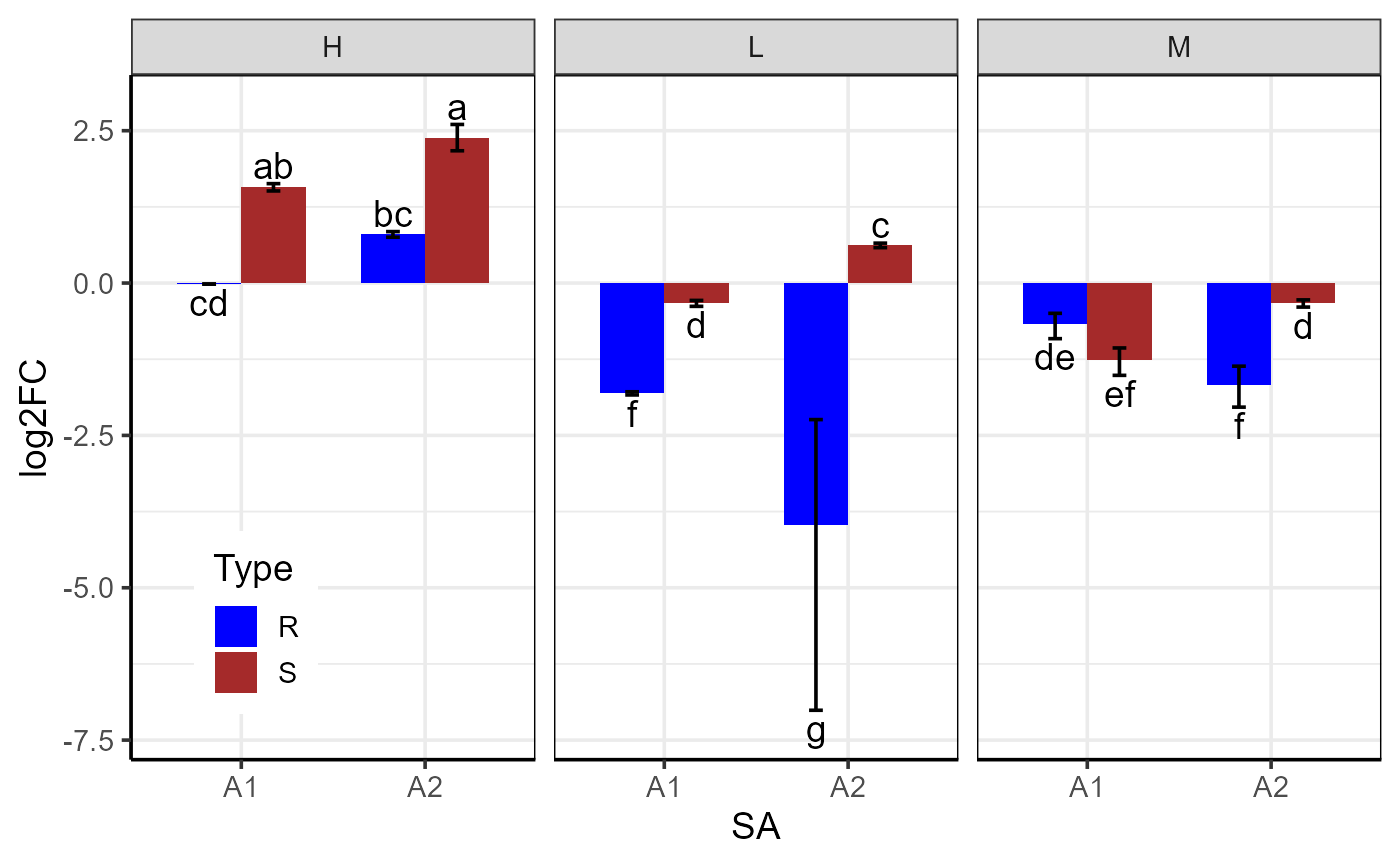
# Generate three-factor bar plot
p <- plotFactor(
df,
x_col = "SA",
y_col = "log2FC",
group_col = "Type",
facet_col = "Conc",
Lower.se_col = "Lower.se.log2FC",
Upper.se_col = "Upper.se.log2FC",
letters_col = "sig",
letters_d = 0.3,
col_width = 0.7,
dodge_width = 0.7,
fill_colors = c("blue", "brown"),
base_size = 14,
alpha = 1,
legend_position = c(0.1, 0.2))
p
data <- read.csv(system.file("extdata", "data_3factor.csv", package = "rtpcr"))
#Perform analysis first
res <- ANOVA_DCt(
data,
numOfFactors = 3,
numberOfrefGenes = 1,
block = NULL)
#> NULL
#>
#> Relative expression (DCt method)
#> Type Conc SA RE log2FC LCL UCL se Lower.se.RE Upper.se.RE
#> 1 S H A2 5.1934 2.3767 8.1197 3.3217 0.1309 4.7428 5.6867
#> 2 S H A1 2.9690 1.5700 4.6420 1.8990 0.0551 2.8578 3.0846
#> 3 R H A2 1.7371 0.7967 2.7159 1.1110 0.0837 1.6391 1.8409
#> 4 S L A2 1.5333 0.6167 2.3973 0.9807 0.0865 1.4441 1.6280
#> 5 R H A1 0.9885 -0.0167 1.5455 0.6323 0.0841 0.9325 1.0479
#> 6 S L A1 0.7955 -0.3300 1.2438 0.5088 0.2128 0.6864 0.9220
#> 7 S M A2 0.7955 -0.3300 1.2438 0.5088 0.2571 0.6657 0.9507
#> 8 R M A1 0.6271 -0.6733 0.9804 0.4011 0.4388 0.4626 0.8500
#> 9 S M A1 0.4147 -1.2700 0.6483 0.2652 0.2540 0.3477 0.4945
#> 10 R M A2 0.3150 -1.6667 0.4925 0.2015 0.2890 0.2578 0.3848
#> 11 R L A1 0.2852 -1.8100 0.4459 0.1824 0.0208 0.2811 0.2893
#> 12 R L A2 0.0641 -3.9633 0.1002 0.0410 0.8228 0.0362 0.1134
#> Lower.se.log2FC Upper.se.log2FC sig
#> 1 2.1705 2.6025 a
#> 2 1.5112 1.6311 ab
#> 3 0.7517 0.8443 bc
#> 4 0.5808 0.6548 c
#> 5 -0.0177 -0.0157 cd
#> 6 -0.3825 -0.2847 d
#> 7 -0.3944 -0.2761 d
#> 8 -0.9127 -0.4968 de
#> 9 -1.5145 -1.0650 ef
#> 10 -2.0363 -1.3641 f
#> 11 -1.8363 -1.7841 f
#> 12 -7.0103 -2.2407 g
#>
#> Combined Expression Table (all genes)
#> gene Type Conc SA RE log2FC LCL UCL se Lower.se.RE
#> 1 PO S H A2 5.1934 2.3767 8.1197 3.3217 0.1309 4.7428
#> 2 PO S H A1 2.9690 1.5700 4.6420 1.8990 0.0551 2.8578
#> 3 PO R H A2 1.7371 0.7967 2.7159 1.1110 0.0837 1.6391
#> 4 PO S L A2 1.5333 0.6167 2.3973 0.9807 0.0865 1.4441
#> 5 PO R H A1 0.9885 -0.0167 1.5455 0.6323 0.0841 0.9325
#> 6 PO S L A1 0.7955 -0.3300 1.2438 0.5088 0.2128 0.6864
#> 7 PO S M A2 0.7955 -0.3300 1.2438 0.5088 0.2571 0.6657
#> 8 PO R M A1 0.6271 -0.6733 0.9804 0.4011 0.4388 0.4626
#> 9 PO S M A1 0.4147 -1.2700 0.6483 0.2652 0.2540 0.3477
#> 10 PO R M A2 0.3150 -1.6667 0.4925 0.2015 0.2890 0.2578
#> 11 PO R L A1 0.2852 -1.8100 0.4459 0.1824 0.0208 0.2811
#> 12 PO R L A2 0.0641 -3.9633 0.1002 0.0410 0.8228 0.0362
#> Upper.se.RE Lower.se.log2FC Upper.se.log2FC sig
#> 1 5.6867 2.1705 2.6025 a
#> 2 3.0846 1.5112 1.6311 ab
#> 3 1.8409 0.7517 0.8443 bc
#> 4 1.6280 0.5808 0.6548 c
#> 5 1.0479 -0.0177 -0.0157 cd
#> 6 0.9220 -0.3825 -0.2847 d
#> 7 0.9507 -0.3944 -0.2761 d
#> 8 0.8500 -0.9127 -0.4968 de
#> 9 0.4945 -1.5145 -1.0650 ef
#> 10 0.3848 -2.0363 -1.3641 f
#> 11 0.2893 -1.8363 -1.7841 f
#> 12 0.1134 -7.0103 -2.2407 g
df <- res$combinedResults
df
#> gene Type Conc SA RE log2FC LCL UCL se Lower.se.RE
#> 1 PO S H A2 5.1934 2.3767 8.1197 3.3217 0.1309 4.7428
#> 2 PO S H A1 2.9690 1.5700 4.6420 1.8990 0.0551 2.8578
#> 3 PO R H A2 1.7371 0.7967 2.7159 1.1110 0.0837 1.6391
#> 4 PO S L A2 1.5333 0.6167 2.3973 0.9807 0.0865 1.4441
#> 5 PO R H A1 0.9885 -0.0167 1.5455 0.6323 0.0841 0.9325
#> 6 PO S L A1 0.7955 -0.3300 1.2438 0.5088 0.2128 0.6864
#> 7 PO S M A2 0.7955 -0.3300 1.2438 0.5088 0.2571 0.6657
#> 8 PO R M A1 0.6271 -0.6733 0.9804 0.4011 0.4388 0.4626
#> 9 PO S M A1 0.4147 -1.2700 0.6483 0.2652 0.2540 0.3477
#> 10 PO R M A2 0.3150 -1.6667 0.4925 0.2015 0.2890 0.2578
#> 11 PO R L A1 0.2852 -1.8100 0.4459 0.1824 0.0208 0.2811
#> 12 PO R L A2 0.0641 -3.9633 0.1002 0.0410 0.8228 0.0362
#> Upper.se.RE Lower.se.log2FC Upper.se.log2FC sig
#> 1 5.6867 2.1705 2.6025 a
#> 2 3.0846 1.5112 1.6311 ab
#> 3 1.8409 0.7517 0.8443 bc
#> 4 1.6280 0.5808 0.6548 c
#> 5 1.0479 -0.0177 -0.0157 cd
#> 6 0.9220 -0.3825 -0.2847 d
#> 7 0.9507 -0.3944 -0.2761 d
#> 8 0.8500 -0.9127 -0.4968 de
#> 9 0.4945 -1.5145 -1.0650 ef
#> 10 0.3848 -2.0363 -1.3641 f
#> 11 0.2893 -1.8363 -1.7841 f
#> 12 0.1134 -7.0103 -2.2407 g
# Generate three-factor bar plot
p <- plotFactor(
df,
x_col = "SA",
y_col = "log2FC",
group_col = "Type",
facet_col = "Conc",
Lower.se_col = "Lower.se.log2FC",
Upper.se_col = "Upper.se.log2FC",
letters_col = "sig",
letters_d = 0.3,
col_width = 0.7,
dodge_width = 0.7,
fill_colors = c("blue", "brown"),
base_size = 14,
alpha = 1,
legend_position = c(0.1, 0.2))
p
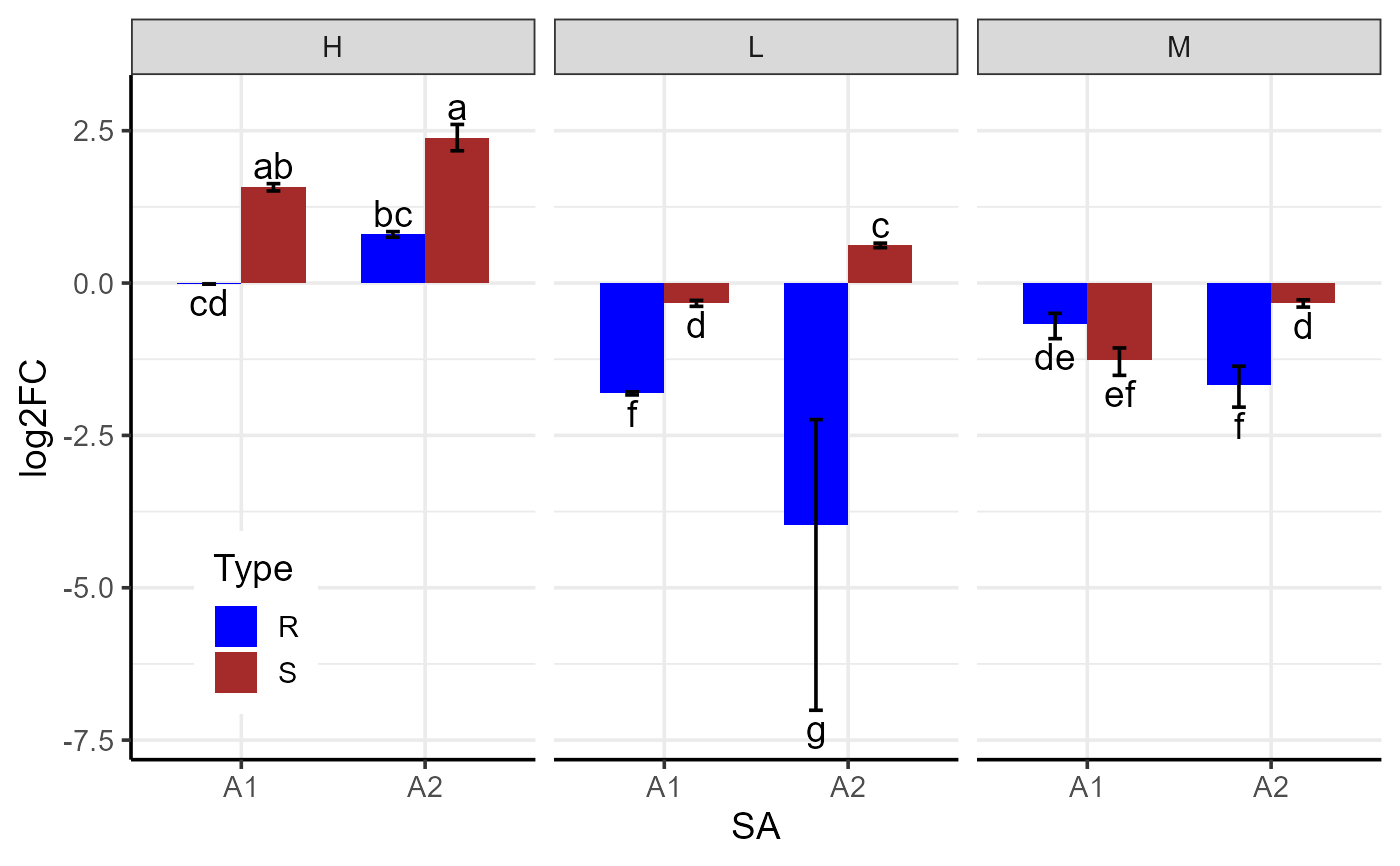 library(ggplot2)
#> Warning: package 'ggplot2' was built under R version 4.4.3
p + theme(
panel.border = element_rect(color = "black", linewidth = 0.5))
library(ggplot2)
#> Warning: package 'ggplot2' was built under R version 4.4.3
p + theme(
panel.border = element_rect(color = "black", linewidth = 0.5))